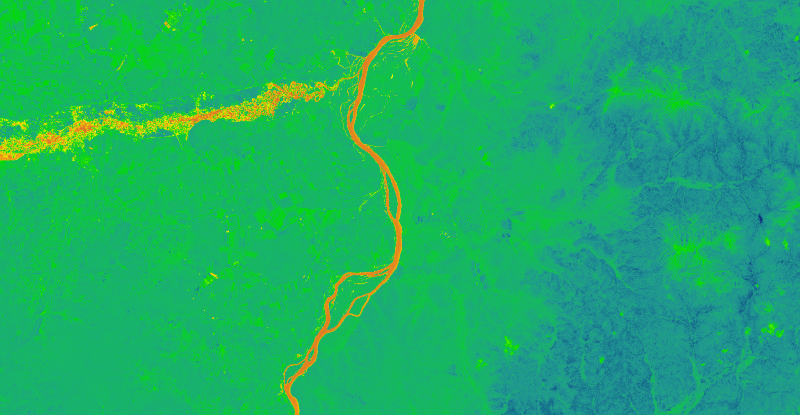
Rasterio package introduction and installation
The Rasterio package, which can read and write GeoTIFF and other formats and return Numpy N-dimensional arrays or GeoJSON geographic json, is a powerful python package for raster data processing developed on GDAL.
Anaconda installs the package with a single command.
If the above command reports an error in the installation, try any of the following lines of the official conda channel installation.
1
2
3
4
5
6
|
conda install -c conda-forge rasterio
conda install -c conda-forge/label/cf202003 rasterio
conda install -c conda-forge/label/dev rasterio
conda install -c conda-forge/label/broken rasterio
conda install -c conda-forge/label/rasterio_dev rasterio
conda install -c conda-forge/label/cf201901 rasterio
|
Other uses can see the officialDOCS
Calculate NDVI code
First import the required packages
1
2
3
4
5
6
|
import rasterio
from rasterio import plot
import matplotlib.pyplot as plt
import numpy as np
import os
from rasterio import transform
|
After the Landsat data is pre-processed and placed in the same folder, the output is only geotiff single image because I am using Python for batch radiometric calibration and 6s atmospheric correction, after placing them all in the same folder, the data to be calculated is as follows
Under each folder, the required band files are parsed, where a total of three bands, B4, B5, and B3, are required to calculate the physical parameters of the plant.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
|
filepath = "D:/Data/Landsat_8_retry_cal_6s"
productIOs = []
pathlist = os.listdir(filepath) #Iterates through the names of the folders in the entire folder and returns a list
#print(pathlist)
for i in pathlist:
product_path = filepath + '/' + str(i)
product_list = os.listdir(product_path)
for product in product_list:
productIOs.append(product.split(".")[0])
for productIO in productIOs:
if productIO[-2:]=='B4':
band4path = product_path + '/' + str(productIO) + '.TIF'
for productIO in productIOs:
if productIO[-2:]=='B5':
band5path = product_path + '/' + str(productIO) + '.TIF'
for productIO in productIOs:
if productIO[-2:]=='B3':
band3path = product_path + '/' + str(productIO) + '.TIF'
band4 = rasterio.open(band4path)
band5 = rasterio.open(band5path)
band3 = rasterio.open(band3path)
red = band4.read(1).astype('float64')
nir = band5.read(1).astype('float64')
green = band3.read(1).astype('float64')
|
After reading the above three bands into the numpy array, you can directly use python mathematical expressions to calculate each vegetation index, the code is as follows.
1
2
3
4
5
6
|
#Vegetation index calculation
ndvi =( nir - red )/(nir+red)
fapar = 107.5*ndvi - 8.0
lai = 5*np.log(ndvi)+ 4.22
ci = nir/green - 1
ccc = ci*0.89+7.47
|
Finally, there is batch output. The results of each of the five calculations are output to the specified folder and saved as geotiff format. In fact, the output of several vegetation indices are written in the same way, just repeat a few times, and will be changed to function form sometime, which can be called directly.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
|
#ndvi
ndvi_output_path = "D:/Data/NDVI_files"
output_ndvi = ndvi_output_path + '/' + str(i) + '_ndvi' +'.TIF'
ndviImage = rasterio.open(output_ndvi,'w',driver='Gtiff',width=band4.width,height = band4.height,\
count=1,crs = band4.crs,transform=band4.transform,dtype='float64')
ndviImage.write(ndvi,1)
ndviImage.close()
#FAPAR
fapar_output_path = "D:/Data/FAPAR_files"
output_fapar = fapar_output_path + '/' + str(i) + '_fapar' +'.TIF'
faparImage = rasterio.open(output_fapar,'w',driver='Gtiff',width=band4.width,height = band4.height,\
count=1,crs = band4.crs,transform=band4.transform,dtype='float64')
faparImage.write(fapar,1)
faparImage.close()
#LAI
LAI_output_path = "D:/Data/LAI_files"
output_LAI = LAI_output_path + '/' + str(i) + '_LAI' +'.TIF'
LAIImage = rasterio.open(output_LAI,'w',driver='Gtiff',width=band4.width,height = band4.height,\
count=1,crs = band4.crs,transform=band4.transform,dtype='float64')
LAIImage.write(lai,1)
LAIImage.close()
#CCC
CCC_output_path = "D:/Data/CCC_files"
output_CCC = CCC_output_path + '/' + str(i) +'.TIF'
CCCImage = rasterio.open(output_CCC,'w',driver='Gtiff',width=band4.width,height = band4.height,count=1,crs = band4.crs,transform=band4.transform,dtype='float64')
CCCImage.write(ccc,1)
CCCImage.close()
|
All codes
Here is the complete code project, written a long time ago, some places can be completely encapsulated as a function, but at that time there is no such awareness, as a learning record it.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
|
import rasterio
from rasterio import plot
import matplotlib.pyplot as plt
import numpy as np
import os
from rasterio import transform
filepath = "D:/Data/Landsat_8_retry_cal_6s"
productIOs = []
pathlist = os.listdir(filepath) #Iterates through the names of the folders in the entire folder and returns a list
#print(pathlist)
for i in pathlist:
product_path = filepath + '/' + str(i)
product_list = os.listdir(product_path)
for product in product_list:
productIOs.append(product.split(".")[0])
for productIO in productIOs:
if productIO[-2:]=='B4':
band4path = product_path + '/' + str(productIO) + '.TIF'
for productIO in productIOs:
if productIO[-2:]=='B5':
band5path = product_path + '/' + str(productIO) + '.TIF'
for productIO in productIOs:
if productIO[-2:]=='B3':
band3path = product_path + '/' + str(productIO) + '.TIF'
band4 = rasterio.open(band4path)
band5 = rasterio.open(band5path)
band3 = rasterio.open(band3path)
red = band4.read(1).astype('float64')
nir = band5.read(1).astype('float64')
green = band3.read(1).astype('float64')
#Vegetation physiological parameters
ndvi =( nir - red )/(nir+red)
fapar = 107.5*ndvi - 8.0
lai = 5*np.log(ndvi)+ 4.22
ci = nir/green - 1
ccc = ci*0.89+7.47
#ndvi
ndvi_output_path = "D:/Data/NDVI_files"
output_ndvi = ndvi_output_path + '/' + str(i) + '_ndvi' +'.TIF'
ndviImage = rasterio.open(output_ndvi,'w',driver='Gtiff',width=band4.width,height = band4.height,\
count=1,crs = band4.crs,transform=band4.transform,dtype='float64')
ndviImage.write(ndvi,1)
ndviImage.close()
#FAPAR
fapar_output_path = "D:/Data/FAPAR_files"
output_fapar = fapar_output_path + '/' + str(i) + '_fapar' +'.TIF'
faparImage = rasterio.open(output_fapar,'w',driver='Gtiff',width=band4.width,height = band4.height,\
count=1,crs = band4.crs,transform=band4.transform,dtype='float64')
faparImage.write(fapar,1)
faparImage.close()
#LAI
LAI_output_path = "D:/Data/LAI_files"
output_LAI = LAI_output_path + '/' + str(i) + '_LAI' +'.TIF'
LAIImage = rasterio.open(output_LAI,'w',driver='Gtiff',width=band4.width,height = band4.height,\
count=1,crs = band4.crs,transform=band4.transform,dtype='float64')
LAIImage.write(lai,1)
LAIImage.close()
#CCC
CCC_output_path = "D:/Data/CCC_files"
output_CCC = CCC_output_path + '/' + str(i) +'.TIF'
CCCImage = rasterio.open(output_CCC,'w',driver='Gtiff',width=band4.width,height = band4.height,count=1,crs = band4.crs,transform=band4.transform,dtype='float64')
CCCImage.write(ccc,1)
CCCImage.close()
|
Appendix
Reference article.
- Rasterio: access to geospatial raster data
- python 栅格处理利器之Rasterio——郭晓隐
- S2ToolBox Level 2 products:LAI, FAPAR, FCOVER